Powering the green plant
SSF reseracher Alexey Amunts is newly published in Nature Plants. Read about the excellent research here!
Chlororibosome is responsible for the synthesis of the energy-making components driving the oxygenic photosynthesis. Amunts and colleagues determined a cryo-EM structure of the chlororibosome with two translational factors. The work reveals unexpected insights into this central protein synthesis machinery in plants and provides a new perspective for the investigation of the evolution of translation and its regulation.
Oxygenic photosynthesis builds a variety of organic compounds, changing the chemistry of the air, the sea and fueling the food chain on our planet. Chemical reactions underpinning this process in plants are taking place in the chloroplast. The chloroplast has its own genome encoding at least 44 proteins, including the central components of the thylakoid membrane that carry out the essential process of oxygenic photosynthesis. These photosynthetic membrane proteins are synthesized by set of dedicated chlororibosomes. Because many of the proteins also coordinate chlorophylls for the light harvesting reactions, the activity of chlororibosomes is likely to be spatiotemporally coupled to the synthesis and incorporation of pigments. This suggests chloroplast-specific regulatory mechanisms and structural adaptation of the chlororibosome. Amunts and colleagues set up to investigate these adaptations and the molecular mechanism of protein synthesis by chlororibosome.
Annemarie Perez Boerema, a PhD student in the lab, purified chlororibosomes from spinach leaves and subjected it to the analysis by electron cryo-microscopy (cryo-EM). The density maps were calculated to 2.91 Å and 3.07 Å for the large and small chlororibosomal subunit respectively. The model of the chlororibosome was then built collectively by all the group members, spearheaded by Shintaro Aibara. Remarkably, the quality of the density map allowed identification of errors in sequencing and mis-annotation in the UniProt database. Particularly, sequences were corrected for eight proteins, and at least six others were re-annotated based on the accurate identification from the density map. In addition, two native translation factors were identified in complex with the chlororibosome: the recycling factor and long hibernation-promoting factor (PSRP1). Both were resolved in the cryo-EM map better than in the corresponding crystal structures of reconstituted ribosomal complexes from E. coli and T. thermophilus.
Figure 1. Native translation factors bound to the chlororibosome.
(A) Comparison of the chloroplast recycling factor density with the X-ray crystallography data for bacterial counterparts, showing a better resolved domain II that allowed its complete building and refinement. (B) The movement of the chloroplast recycling factor domain II (left) is caused by the interactions with proteins bL27c and PSRP1 (right).
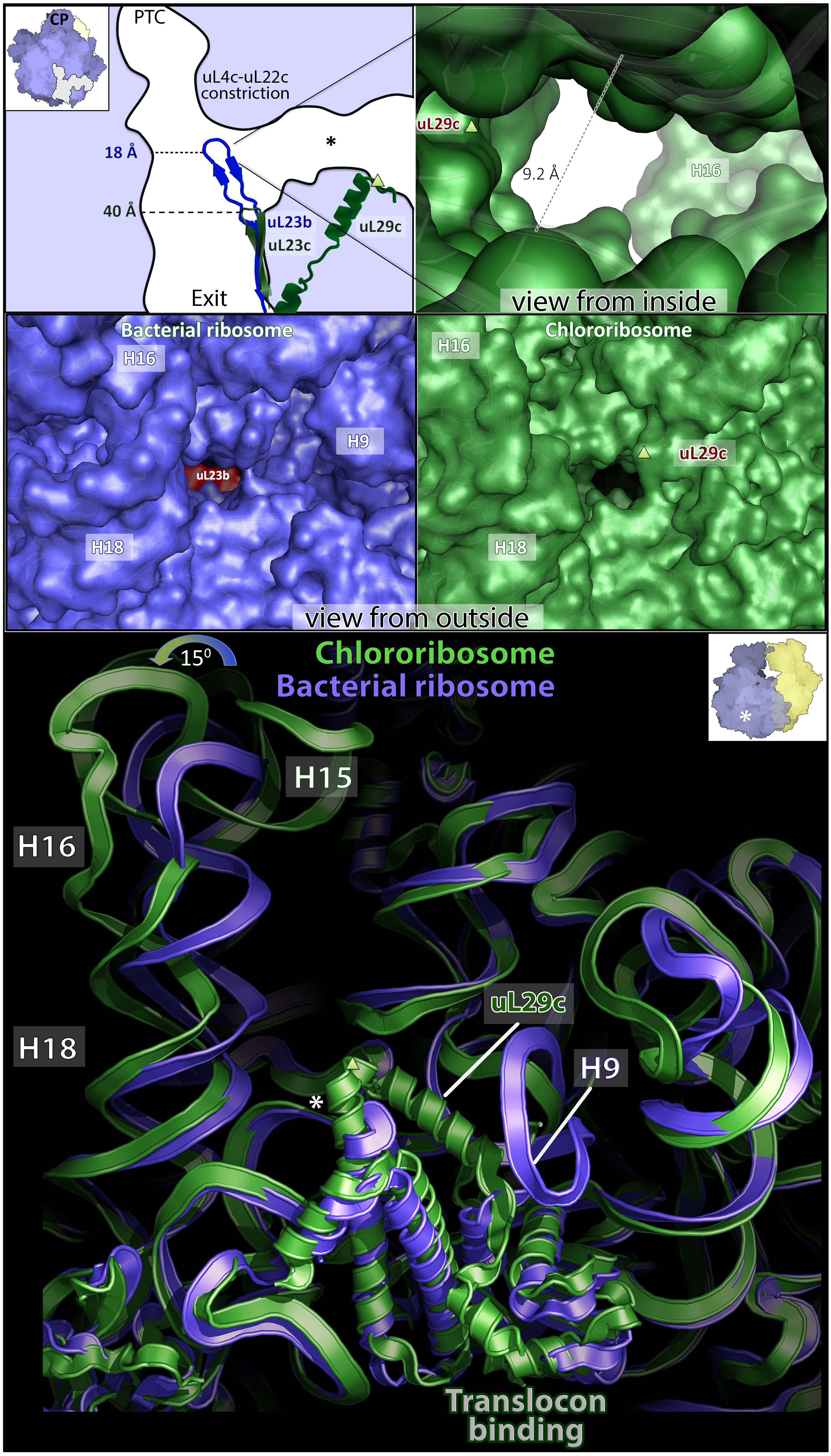
The model of the spinach chlororibosome reveals several molecular specialties of the large subunit. The most intriguing is a prominent channel, at least 9 Å wide X 30 Å in length, extending from the exit tunnel to the chlororibosome exterior. In addition, structural re-arrangements on the surface led to an increased area for a translocon binding. Finally, protein deletion resulted in more than twofold widening of the tunnel that is likely to affect an initial protein folding.
Figure 2. Remodeling of the large subunit.
Top panel, shortening of the chloroplast hairpin loop of uL23c (green) as compared to bacterial uL23b (blue) results in the new channel (asterisk). The two exits are connected through uL29c. Middle panel, view from outside shows that the channel does not exist in bacterial ribosome. Bottom panel, chl-rRNA rearrangement at the exit from the new channel. uL29c extends from the exit towards a putative translocon binding site. Asterisk and triangle indicate reference points between the panels.
When Amunts and colleagues analyzed how the large subunit is coordinated with the small subunit, they noticed that a protein-protein intersubunit bridge that in the beginning appeared to be specific to the chlororibosome turned out to be similar to the one found in an unrelated ribosome from human mitochondria. This prompted the authors to assess if there might be additional means of structural adaptations taken parallel evolutionary paths in chloro- and mitoribosomes. They found such correlations on the level of rRNA sequence variability, as well as on the protein level, in particular the association of the protein bS1c with the small subunit in a way that does not allow the long hibernation-promoting factor to dimerize.
These unexpected insights into the light-driven photosynthetic protein synthesis machinery in plants are central to biology and provide a new perspective for the investigation of the evolution of translation and its regulation.
Further references